Study: Novel algorithm 6 times more reliable at predicting defective RNA
So far, it has not been possible to explain the causes of around half of all rare hereditary diseases. A Munich research team has developed an algorithm that predicts the effects of genetic mutations on RNA formation six times more precisely than previous models. As a result, the genetic causes of rare hereditary diseases and cancer can be identified more precisely.
Variations of genetic sequence occur relatively frequently—on average, one in a thousand nucleotide of a person’s genome is affected. In rare cases, these changes can lead to defective RNAs and hence non-functional proteins.
This can lead to dysfunction in individual organs. If a rare disease is suspected, computer-assisted diagnosis programs can help in the search for possible genetic causes. Specifically the genome can be analyzed using algorithms to find out whether there is a connection between rare genetic variations and dysfunctions in specific parts of the body.
Interdisciplinary research project
Under the leadership of Julien Gagneur, Professor of Computational Molecular Medicine at the Technical University of Munich (TUM) and leader of the Computational Molecular Medicine research group at Helmholtz Munich, an interdisciplinary team from the Informatics and Medicine departments developed a new model that is better than its predecessors at predicting which DNA variations will lead to incorrectly formed RNA.
“A reliable diagnosis can be made for about half of our patients using established DNA analysis methods,” says Dr. Holger Prokisch, co-author of the study and group leader of the Institute of Human Genetics at TUM and Helmholtz Munich. “For the rest, we need models that improve our predictions. Our newly developed algorithm can make an important contribution to this.”
Focus of the model is on splicing
In their study, the researchers considered genetic variations that influence the conversion process of DNA into RNA and ultimately the formation of proteins in a tissue-specific fashion.
The focus was on splicing—a process in the cells where the RNA is cut in such a way that the building instructions for the protein can be read later. If there is variation on the DNA, this process can be disrupted and results in either too much or too little being cut from the RNA. Errors in the splicing process are thought to be one of the most common causes of incorrect protein formation and hereditary diseases.
Significantly greater precision than previous studies
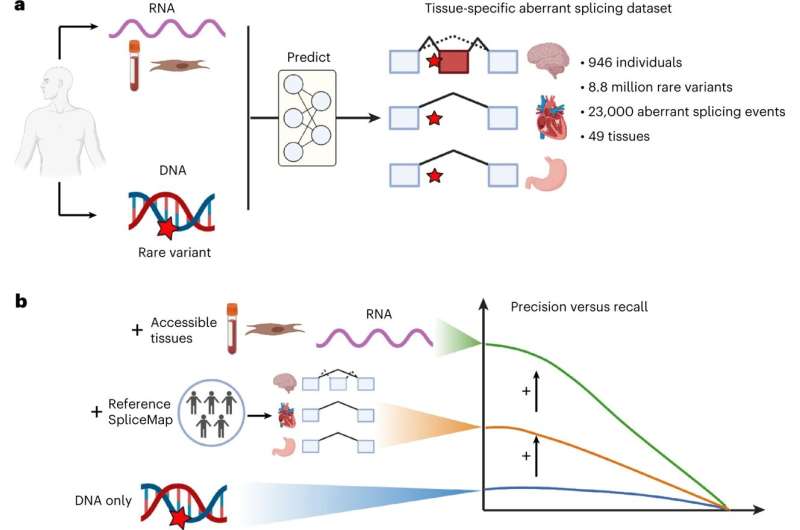
The team leverage on existing data sets in order to be able to make statements about possible associations between genetic variations and splicing dysfunctions in specific tissues. These data sets contain DNA and RNA samples from 49 tissues from a total of 946 individuals.
In comparison to previous studies, the team initially considered each sample to see if and to what extent incorrect splicing resulting from variation on the DNA generally manifests itself through splicing dysfunctions in certain tissues. For example, a protein may be relevant for special areas of the heart, while it may have no function in the brain.
“For this purpose, we created a tissue-specific splicing map in which we quantified which places on the RNA are important to splicing in a given tissue. Thanks to our approach, we were able to limit our model to biologically relevant contexts. The skin and blood samples we used enabled us to draw conclusions about hard-to-reach tissues, such as the brain or the heart,” says Nils Wagner, lead author of the study and doctoral student at the Chair of Computational Molecular Medicine at TUM.
In the analysis, each gene with at least one rare genetic variant and that is relevant for protein formation was considered. In addition to the protein-coding sections on the RNA, there are sections that are important for other processes in our cells. These were not considered in the study. This resulted in a total of nearly 9 million rare genetic variants being studied.
“Thanks to our newly developed model, we were able to increase the precision of predicting incorrect splicing sixfold in comparison to previous models. At a recall of 20 percent, previous algorithms achieved a precision of 10 percent. Our model achieves a precision of 60 percent at the same recall,” says Prof. Julien Gagneur.
Precision and recall are important metrics for projecting the effectiveness of models. The precision indicates how many of the genetic variations predicted by the model actually lead to incorrect splicing. The recall shows how many genetic variations mutations that lead to incorrect splicing are recovered by the model.
“We achieved such a large advance in precision by looking at the splicing process in a tissue-specific way and by using direct splicing measurements from easily accessible tissues such as blood or skin cells in order to predict splicing errors in inaccessible tissues like the heart or the brain,” says Prof. Julien Gagneur.

Practical use of the algorithm
The model is being used as part of the European research project “Solve-RD—solving the unsolved rare diseases.” The initiative has set itself the goal of improving the diagnostic outcomes for rare diseases through a broad exchange of knowledge. The team from TUM has already analyzed 20,000 DNA sequences from a total of 6,000 affected families.
Furthermore, the model should make it possible to more easily find the genetic diagnosis of various forms of leukemia in the future. For this purpose, researchers are currently examining 4,200 DNA and RNA samples from leukemia patients.
The work is published in the journal Nature Genetics.
More information:
Nils Wagner et al, Aberrant splicing prediction across human tissues, Nature Genetics (2023). DOI: 10.1038/s41588-023-01373-3
Journal information:
Nature Genetics
Source: Read Full Article