The rollout of vaccines is on in an attempt to achieve population immunity against the coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, the recent emergence of several variants that have altered transmissibility, infectivity and virulence characteristics has cast doubt on the feasibility of this goal.
A new preprint, recently released on the bioRxiv* server, explores the impact of a single mutation, the P681H mutation, on the transmissibility of the virus.
.jpg)
Two SARS-CoV-2 lineages are now in circulation, both stemming from the original or wildtype Wuhan-Hu1 strain. Lineage B, especially the D614G-mutant strains, have since become dominant the world over as this mutation stabilizes the critical spike protein and thus enhances viral transmissibility.
The virus depends on its spike protein to engage the target host cell receptor, the angiotensin-converting enzyme 2 (ACE2), and achieve cell entry. The spike has undergone a number of point and grouped mutations.
Towards the end of 2020, the so-called UK variant of SARS-CoV-2 was detected as it led to failure to detect the target spike gene in spike-based diagnostic polymerase chain reaction tests, though the subjects were reported positive by PCR tests detecting other viral protein targets like the nucleocapsid.
Sequencing led to its being termed variant of concern (VOC) B.1.1.7/501Y.V1, and it rapidly spread over the south of England and then to the rest of the UK and the world. Interestingly, this VOC has 23 point mutations in its genome, especially nine in the spike protein.
These include a 69-70 deletion (the cause of S-gene target failure), Y144 del, N501Y, A570D, D614G, P681H, T716I, S982A, and D118H. Seven of these are found only in the UK strain.
The D614G and jN501Y mutations are found in all the three VOCs currently circulating, namely, the UK, South African (B.1.351) and Brazil (B.1.1.28.1 (P.1), variants, respectively. Both these mutations enhance the binding affinity of the spike to the ACE2 receptor.
While the UK variant has higher transmissibility, and perhaps an association with greater severity of disease, it appears to be neutralized by the same antibodies as the wildtype spike. It may be that, like the influenza virus, “the B.1.1.7 variant acquired its extensive range of mutations in a single immunocompromised individual who then initiated a super-spreader event that gave rise to the subsequent dissemination of the lineage.”
The furin cleavage site
The virus spike is made up of S1 and S2 subunits, at the interface of which there is a furin cleavage site. This is not found in other Sarbecoviruses.
This interface in SARS-CoV-2 shows the sequence 681-P-R-R-A-R|S-686, and cleavage occurs between the arginine and serine residues, termed the P1|P1’ residues. In this case, the P681H mutation in the UK strain occurs at the P5 cleavage position.
Furin is a serine protease that requires a polybasic motif-containing paired arginine residues, at least R-xx-R (P4-x-x-P1), but prefers that the P2 also be basic. The presence of a furin cleavage site in SARS-CoV-2 makes it more transmissible.
This site is not a typical cleavage site for furin, however. With the UK variant, the H at P5 may result in an additional basic residue, particularly at low pH, therefore altering its susceptibility to furin cleavage and its infectivity.
Nonetheless, this introduction still does not make it a consensual furin cleavage site.
Study aims
The study aimed to understand how proteolytic activity was involved in spike-mediated infection with the UK variant, especially with the P681H mutation in the spike
What were the results?
First using bioinformatics tools, the researchers found that the UK spike variant was predicted to have slightly greater furin cleavage compared to the wildtype spike, while no cleavage was expected for the SARS-CoV and low cleavage for the MERS-CoV (Middle East respiratory syndrome coronavirus). The seasonal endemic coronaviruses showed much higher cleavage scores.
Biochemical assays of peptide cleavage showed that compared to trypsin, furin showed effective cleavage of the wildtype S1/S2 interface, and at pH 7.5 the cleavage increased slightly for the UK variant.
At pH 7 and 6.5, however, the cleavage actually dropped, and below 6.5 it was no longer measurable. Thus, the P681H does not significantly enhance proteolytic cleavage at the interface and may even inhibit it.
In the cell-based assays, both early cell entry pathways, using Vero-TMPRSS2-expressing cells, and late entry, using Vero E6 cells, were tested. In the former, the virus S2’ priming site is activated by the host serine protease TMPRSS2, and the latter by cathepsin L.
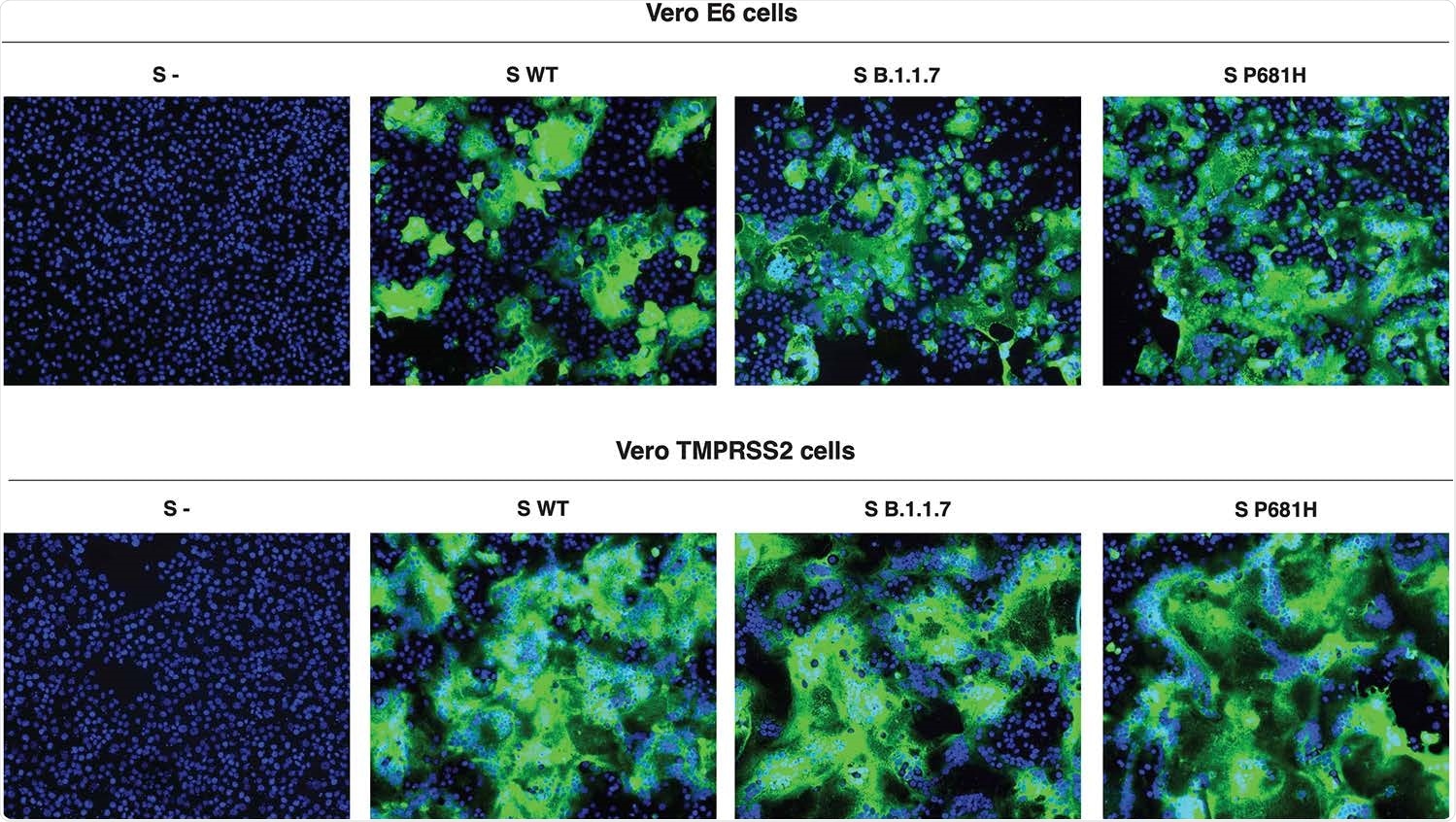
The former cell line was more susceptible to SARS-CoV-2 entry, but the various SARS-CoV-2 spike-expressing pseudoparticles accomplished comparable levels of entry. However, the spike cleavage was markedly increased with the two spike variants containing the P681H mutation.
The ratios of cleaved: uncleaved spike observed with the UK and P681H-mutant wildtype virus were increased three-fold and over five-fold, respectively, relative to the wildtype virus. Further studies will show if this is mediated by furin or another protease.
Cell-cell fusion assays, showing membrane fusion activity, showed the Vero-TMPRSS2 cells to be more active at forming syncytia than the Vero E6 cells, but no difference between the spikes themselves, as before.
What are the implications?
The findings suggest that transduction of the spike variants by pseudoparticles is not affected significantly by the presence of the P681H mutation, even though it appears to increase cleavage efficiency.
Cell-cell fusion activity is also not affected. Thus, the only difference is in the extent of spike cleavage by furin-like proteases, but without impacting the entry of the virus into the cells or membrane fusion.
“We consider that other factors are at play to account for the increased in transmission and disease severity attributed to the SARS-CoV-2 B.1.1.7 VOC.” This agrees with an earlier study using the infectious virus itself, indicating no difference in viral replication in primary human airway cells with the UK variant.
Another study showed that in cells with a low level of expression of the ACE2 receptor, the entry of the virus was enhanced.
This mutation has been observed also in B.1.243, which was for a time dominant in New York, eventually replaced by B.1.1.7 and B.1.222 as well as non-P681H mutants. The increased spike cleavage observed in this study may not always be a good thing.
On the other hand, it may signal the change from a pandemic to an endemic respiratory virus such as the seasonal coronaviruses HCoV-HKU1 and HCoV-OC43, which both have polybasic furin cleavage sites at this position. More study will reveal if this is related to increased or prolonged viral shedding, which could explain the higher transmissibility of this variant.
The study thus shows that the epidemiological context remains vital in addition to the analysis of the effect of individual point mutations on viral biology. The UK variant is currently susceptible to already formed antibodies against the wildtype virus, unlike the SA and Brazil VOCs. This could change if the virus picks up E484K or similar mutations during its spread in the community.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Lubinski, B. et al. (2021). Functional evaluation of proteolytic activation for the SARS-CoV-2 variant B.1.1.7: role of the P681H mutation. bioRxiv preprint. doi: https://doi.org/10.1101/2021.04.06.438731, https://www.biorxiv.org/content/10.1101/2021.04.06.438731v1
Posted in: Medical Science News | Medical Research News | Miscellaneous News | Disease/Infection News | Healthcare News
Tags: ACE2, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Arginine, binding affinity, Bioinformatics, Cell, Cell Line, Coronavirus, Diagnostic, Enzyme, Gene, Genome, Influenza, MERS-CoV, Mutation, Pandemic, pH, Polymerase, Polymerase Chain Reaction, Protein, Receptor, Respiratory, SARS, SARS-CoV-2, Serine, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Virus

Written by
Dr. Liji Thomas
Dr. Liji Thomas is an OB-GYN, who graduated from the Government Medical College, University of Calicut, Kerala, in 2001. Liji practiced as a full-time consultant in obstetrics/gynecology in a private hospital for a few years following her graduation. She has counseled hundreds of patients facing issues from pregnancy-related problems and infertility, and has been in charge of over 2,000 deliveries, striving always to achieve a normal delivery rather than operative.
Source: Read Full Article
