Mapping the development of infection-fighting immune cells
The immune system protects the body from invaders, such as bacteria, viruses, or tumors, with its intricate network of proteins, cells, and organs. Specialized immune cells, called cytotoxic T cells, can develop into short-lived effector cells that kill infected or cancerous cells within our bodies.
A small portion of those effector cells remain after an infection and become longer-lived memory cells, which “remember” infections and respond when infections reappear. But little was known about what influences cytotoxic T cells to transform into these effector and memory T cell subtypes.
Now, Salk Institute Professor Susan Kaech and Associate Professor Diana Hargreaves have discovered that a protein complex called cBAF can open or close genetic “doors” to control T cell fate. The study, published in Immunity, illuminates how T cells fight and remember infections and paves the way for the development of more effective vaccines and cancer therapeutics.
“We’ve shown how T cells transform into specialized subtypes during an infection and what happens when they mess up the genetic blueprint. Without this genetic blueprint, T cells may not be able to resolve or remember infections, or could possibly become cancerous, like we see in T cell lymphoma,” says Kaech, senior and co-corresponding author, director of the NOMIS Center for Immunobiology and Microbial Pathogenesis, and holder of the NOMIS Chair.
“Finding that the cBAF protein complex is essential in creating different T cell subtypes opens up new questions, like can we control this process to optimize immune cell function for cancer therapeutics?”
The question of how cytotoxic T cells develop into effector and memory T cells can be answered by looking at how they are created—a process that relies on genetic instructions contained in chromatin. Chromatin is composed of a mixture of genetic code (DNA) that works as instructions, as well as proteins and RNA that regulate and protect those instructions.
Rearranging chromatin within a cell exposes or conceals different sections of genetic code, in turn altering the available instructions. Once instructions are available, proteins called transcription factors determine if and how often they are read.
The researchers knew that chromatin works like scaffolding—protecting the genetic code within and shielding it from transcription factors. For transcription factors to access those instructions, they likely need help from chromatin-remodeling cBAF, which can signal proteins and RNA to reveal genetic code for transcription factors.
“Chromatin remodeling is essential in determining the identities of cells,” says co-first author Bryan McDonald, a graduate student in the Kaech lab. “In other cell types, the chromatin remodeling complex cBAF was known to play a major role in cellular differentiation. We wanted to know if cBAF was important for cytotoxic T cell differentiation, too.”
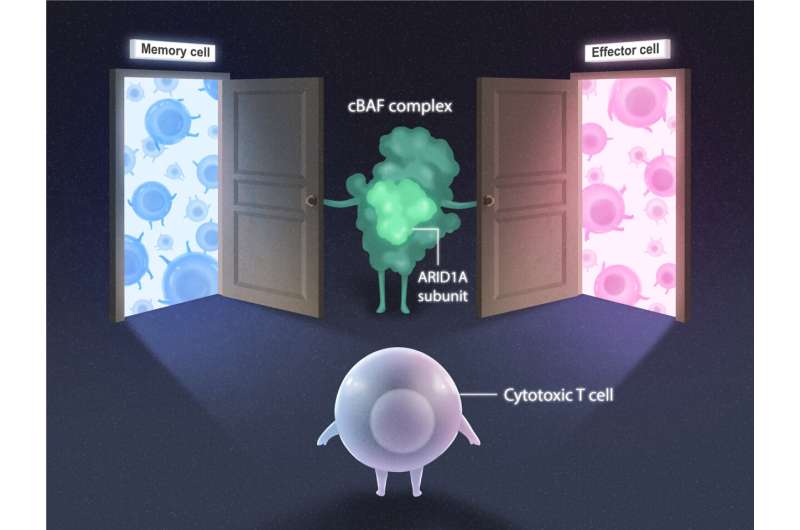
The cBAF complex is made up of many subunits, including ARID1A. To explore the role of ARID1A on chromatin remodeling in T cells, the team examined T cells with and without ARID1A subunits in mice. After a week-long viral infection (to instigate the T cell immune response), the researchers looked for changes in chromatin structure, location of proteins, and genetic code accessibility.
“The chromatin landscape evolved immensely over the course of the infection,” says co-first author Brent Chick, a graduate student in the Hargreaves lab. “cBAF complexes bearing the ARID1A subunit were crucial in rearranging chromatin so transcription factors could access the genetic code that instructs the development of different T cell subtypes with specialized functions.”
Without ARID1A, the cytotoxic T cells could not turn into the short-lived, rapid-response effector subtype. The loss of ARID1A did not prevent the cytotoxic T cells from turning into their longer-lived memory subtype, but the memory cells made without ARID1A were not as effective at responding to a viral reinfection. Finally, the formation of memory T cells that reside in organs, like the lungs or small intestine, was dependent on ARID1A. Without ARID1A, tissue-resident memory T cells could not develop at all.
ARID1A was the key player, as the team suspected. And the mechanism was now clear—during infections, cBAF complexes with ARID1A open different chromatin “doors” to reveal genetic code to transcription factors, which then determine the fate of the cytotoxic T cells. Establishing this novel mechanism paves the way for investigation into how chromatin remodeling, genetic code, and transcription factors together contribute to a successful immune system response.
“Our research demonstrates that removing ARID1A from the immune response equation may greatly damage our ability to fight infection,” says Hargreaves, co-corresponding author and holder of the Richard Heyman and Anne Daigle Endowed Developmental Chair. “I hope that our findings inspire new questions in infectious disease and cancer research—and that we have demonstrated the value of asking those questions collaboratively.”
Next, the scientists plan to examine this mechanism using human T cells. Additionally, they aim to discern how memory T cells are affected by this process, since they are valuable prognostic tools for assessing human tumors.
More information:
Susan M. Kaech, Canonical BAF complex activity shapes the enhancer landscape that licenses CD8+ T cell effector and memory fates, Immunity (2023). DOI: 10.1016/j.immuni.2023.05.005. www.cell.com/immunity/fulltext … 1074-7613(23)00222-4
Journal information:
Immunity
Source: Read Full Article