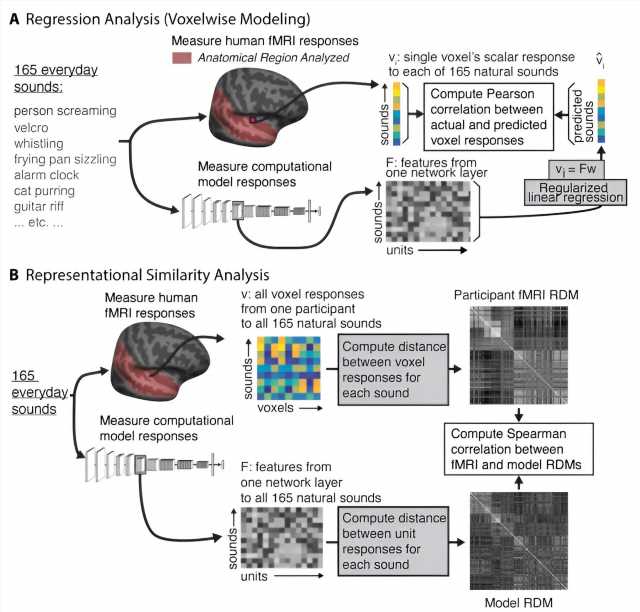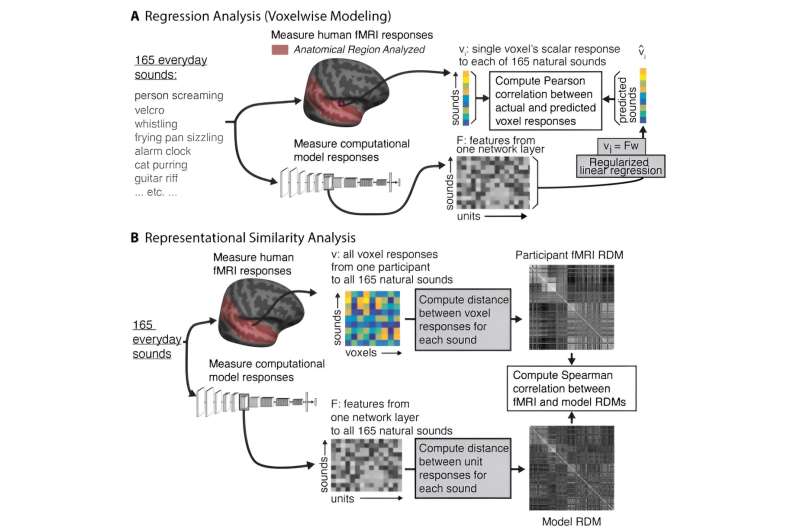
Computational models that mimic the structure and function of the human auditory system could help researchers design better hearing aids, cochlear implants, and brain-machine interfaces. A new study from MIT has found that modern computational models derived from machine learning are moving closer to this goal.
In the largest study yet of deep neural networks that have been trained to perform auditory tasks, the MIT team showed that most of these models generate internal representations that share properties of representations seen in the human brain when people are listening to the same sounds.
The study also offers insight into how to best train this type of model: The researchers found that models trained on auditory input including background noise more closely mimic the activation patterns of the human auditory cortex.
“What sets this study apart is it is the most comprehensive comparison of these kinds of models to the auditory system so far. The study suggests that models that are derived from machine learning are a step in the right direction, and it gives us some clues as to what tends to make them better models of the brain,” says Josh McDermott, an associate professor of brain and cognitive sciences at MIT, a member of MIT’s McGovern Institute for Brain Research and Center for Brains, Minds, and Machines, and the senior author of the study.
MIT graduate student Greta Tuckute and Jenelle Feather Ph.D. ’22 are the lead authors of the open-access paper published in PLOS Biology.
Models of hearing
Deep neural networks are computational models that consists of many layers of information-processing units that can be trained on huge volumes of data to perform specific tasks. This type of model has become widely used in many applications, and neuroscientists have begun to explore the possibility that these systems can also be used to describe how the human brain performs certain tasks.
“These models that are built with machine learning are able to mediate behaviors on a scale that really wasn’t possible with previous types of models, and that has led to interest in whether or not the representations in the models might capture things that are happening in the brain,” Tuckute says.
When a neural network is performing a task, its processing units generate activation patterns in response to each audio input it receives, such as a word or other type of sound. Those model representations of the input can be compared to the activation patterns seen in fMRI brain scans of people listening to the same input.
In 2018, McDermott and then-graduate student Alexander Kell reported that when they trained a neural network to perform auditory tasks (such as recognizing words from an audio signal), the internal representations generated by the model showed similarity to those seen in fMRI scans of people listening to the same sounds.
Since then, these types of models have become widely used, so McDermott’s research group set out to evaluate a larger set of models, to see if the ability to approximate the neural representations seen in the human brain is a general trait of these models.
For this study, the researchers analyzed nine publicly available deep neural network models that had been trained to perform auditory tasks, and they also created 14 models of their own, based on two different architectures. Most of these models were trained to perform a single task—recognizing words, identifying the speaker, recognizing environmental sounds, and identifying musical genre—while two of them were trained to perform multiple tasks.
When the researchers presented these models with natural sounds that had been used as stimuli in human fMRI experiments, they found that the internal model representations tended to exhibit similarity with those generated by the human brain. The models whose representations were most similar to those seen in the brain were models that had been trained on more than one task and had been trained on auditory input that included background noise.
“If you train models in noise, they give better brain predictions than if you don’t, which is intuitively reasonable because a lot of real-world hearing involves hearing in noise, and that’s plausibly something the auditory system is adapted to,” Feather says.
Hierarchical processing
The new study also supports the idea that the human auditory cortex has some degree of hierarchical organization, in which processing is divided into stages that support distinct computational functions. As in the 2018 study, the researchers found that representations generated in earlier stages of the model most closely resemble those seen in the primary auditory cortex, while representations generated in later model stages more closely resemble those generated in brain regions beyond the primary cortex.
Additionally, the researchers found that models that had been trained on different tasks were better at replicating different aspects of audition. For example, models trained on a speech-related task more closely resembled speech-selective areas.
“Even though the model has seen the exact same training data and the architecture is the same, when you optimize for one particular task, you can see that it selectively explains specific tuning properties in the brain,” Tuckute says.
McDermott’s lab now plans to make use of their findings to try to develop models that are even more successful at reproducing human brain responses. In addition to helping scientists learn more about how the brain may be organized, such models could also be used to help develop better hearing aids, cochlear implants, and brain-machine interfaces.
“A goal of our field is to end up with a computer model that can predict brain responses and behavior. We think that if we are successful in reaching that goal, it will open a lot of doors,” McDermott says.
More information:
Greta Tuckute et al, Many but not all deep neural network audio models capture brain responses and exhibit correspondence between model stages and brain regions, PLOS Biology (2023). journals.plos.org/plosbiology/ … journal.pbio.3002366. On bioRxiv: DOI: 10.1101/2022.09.06.506680
Journal information:
PLoS Biology
,
bioRxiv
Source: Read Full Article