Among the things to give thanks for this year is the fact that stubborn, persistent problems seem to have an irresistible sort of magnetic pull for scientists and mathematicians. Things that would leave most of us bewildered are their daily bread.
And that is one reason researchers are making progress in the ongoing fight against cancer, viruses and other diseases.
Such is the case for Abhyudai Singh, a mathematical biologist who is an associate professor in the University of Delaware’s Department of Electrical and Computer Engineering, Mathematical Sciences and Biomedical Engineering and an affiliated faculty member of the Center for Bioinformatics and Computational Biology.
Singh studies how biochemical processes work inside living cells. Specifically, he develops mathematical models that provide insight into the complex, sometimes hidden processes involved in cellular networks and what happens when they go wrong.
His computational work has been a significant part of a collaborative effort with a team at the University of Pennsylvania, led by Professor Arjun Raj of Penn’s Department of Bioengineering. Their recent findings were published in the journal Cell. The article, whose first author was Dr. Sydney M. Shaffer, assistant professor at Penn’s Perelman School of Medicine, shows a potential new strategy against therapy-resistant melanoma cells that could be useful against all kinds of resistant cancers.
“Cancer cells are heterogeneous, even in the same tumor,” Singh said. “Some have drug tolerance and survive therapy. Then they rebound…. Now we have found molecular mechanisms involved in these drug-tolerant cells that have implications for therapy and can be applied to different cancer types.”
The goal was to identify all of the genes that produced drug tolerance in cells, hoping to then map out a way to circumvent their ability to survive these therapies.
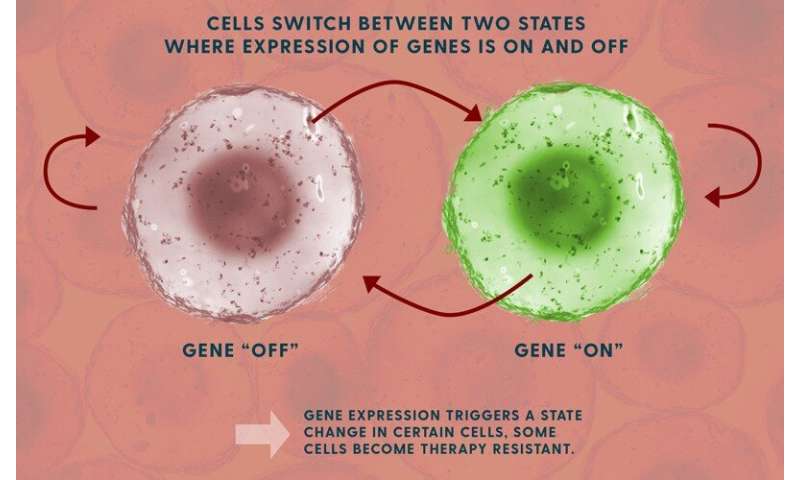
The team studied fluctuations in the ways the cells express their genetic traits, turning them “on” and “off.” This process depends on messenger RNA molecules, which convert the genetic code into specific proteins.
They found that a subset of rare cells carried a biological “memory” that could survive multiple generations of cell division.
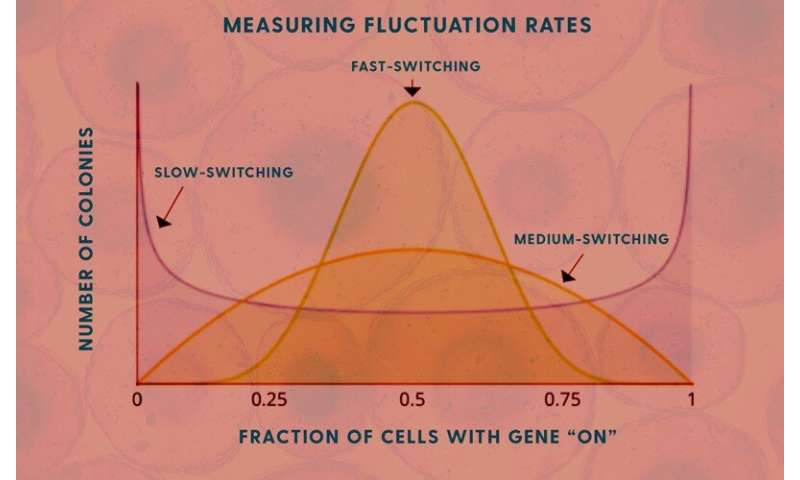
Singh’s model shows how the speed of the cells’ genetic fluctuations is related to levels of messenger RNA in the cells. Knowing those levels allows his model to calculate how fast these genes are turning “on” and “off.”
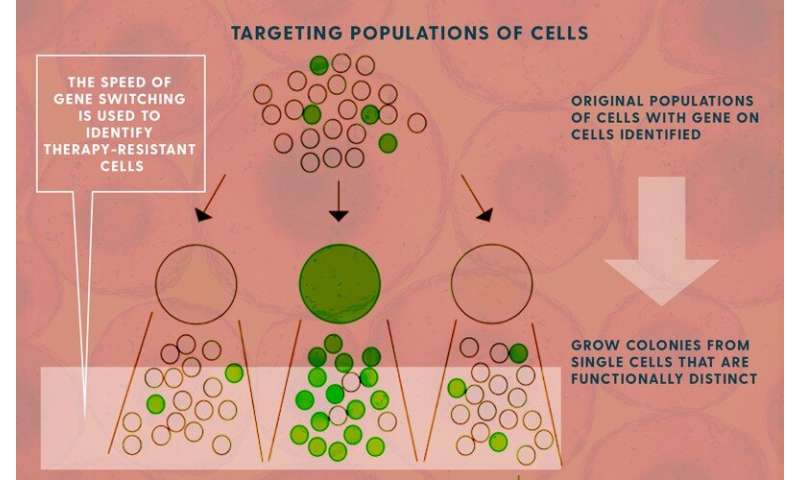
Once you know when those genes turn off—essentially “forgetting” the resistance they had against the drug therapy—you can target new therapies and new therapy schedules to arrive at that vulnerable time.
Different genes have different memories, Singh said. Some are “on” for five or six generations before they turn off, some are on for eight generations. This reflects a “transient heritability” that eventually ends.
Singh and his colleagues used the classic Luria and Delbrück fluctuation analysis, named for 1969 Nobel Prize winners Max Delbrück and Salvador Luria, in this work and used the MemorySeq method to find the levels of on/off genetic fluctuations in these melanoma cells.
“You can get the fluctuation, but does it mean one generation or two or three? That comes from the modeling quite elegantly,” Singh said.
This modeling approach adds a powerful time element that isn’t possible when a drug is added to a sample or genetic sequencing is done, destroying the cell line.
Source: Read Full Article
