Antibody that inhibits broad range of sarbecoviruses found
Scientists have discovered an antibody that may lead to more effective treatments against a wide range of sarbecoviruses, the family of viruses that includes the coronavirus SARS-CoV-2 and their variants.
The antibody neutralizes SARS-CoV-2, SARS-CoV-2 variants and other sarbecoviruses in the lab, protects against infection in animal studies, and appears to be able to thwart the virus’ attempts to evade it through mutation.
“Our findings suggest it’s a very good candidate for clinical development as monoclonal antibody treatment,” said David Veesler, investigator with the Howard Hughes Medical Institute and associate professor of biochemistry at the University of Washington School of Medicine in Seattle, who led the research effort with Matteo Samuele Pizzuto and Davide Corti of Humabs Biomed SA, a subsidiary of Vir Biotechnology.
The researchers report their findings in the journal Science. Young-Jun Park, a senior scientist in the Veesler lab and Anna De Marco of Humabs Biomed were lead authors of the paper, which was published Thursday, Jan. 6.
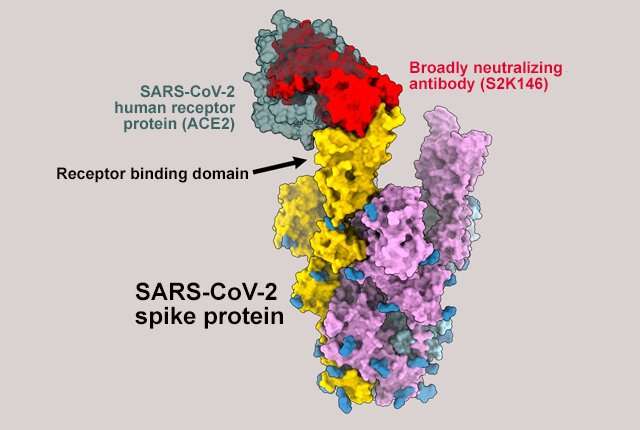
The antibody, designated S2K146, was isolated from the antibody-producing B-cells of a patient who had COVID-19 and recovered. Like other antibodies induced by SARS-CoV-2 vaccines and used in monoclonal antibody treatments, S2K146 targets the viral spike protein.
The spike protein latches onto a protein found on the surface of the cells called angiotensin-converting enzyme (ACE2). It then initiates the process by which the virus enters the cell. The region of the spike protein that binds to ACE2 is called its receptor-binding domain and is the main target of antibodies after infection or vaccination.
One way that antibodies to SARS-CoV-2 prevent infection is by binding to the spike protein receptor-binding domain so it cannot latch onto ACE2. Over time, however, variants, like omicron, acquire mutations that change the amino acid sequences of the spike protein’s receptor binding domain so that it is no longer recognized by many antibodies against it. This is called immune evasion.
For the virus, there’s a potential drawback with this strategy: the changes in the receptor binding domain that allow it to escape the antibody may also impair the ability of the spike protein to bind to its target on the cell, ACE2, and initiate infection.
The SARS-CoV-2 spike protein, however, has proved to be highly adaptable and variants have emerged with mutations that allow the receptor binding domain to escape existing antibodies against it while still being able to bind to ACE2 and trigger infection. Because of the extraordinary mutational “plasticity” of the spike region involved in ACE2 binding, said Veesler, many researchers did not think antibodies targeting it would be broadly neutralizing viruses as different as SARS-CoV-2 and SARS-CoV.
S2K146 appears to be different. The area it uses to bind to and block the spike protein receptor-binding domain is almost identical to the region recognizing the ACE2 receptor. “S2K146 mimics the molecular contacts formed with the ACE2 receptor by binding exactly where the spike protein needs to attach to the cell,” Veesler said.
As a result, any change in the viral spike that reduces the ability of the antibody to bind to it also appears to reduce the ability of the virus to bind to ACE2 and infect cells.
This was borne out in an experiment Veesler and his colleagues did as part of their study. In this experiment, they exposed a surrogate virus that carries the SARS-CoV-2 spike protein to S2K146 to see if escape mutants would emerge. They did this tens of times, and only one escape mutant emerged. But its ability to bind to ACE2 was so poor it could not outcompete the original virus. The finding suggests it will be very difficult, though not impossible, for variants to emerge that can both evade S2K146 and remain fit, Veesler said.
Source: Read Full Article