Antibiotic-Resistant Bacteria and Sewage
Antibiotic resistance is a growing threat to the treatment and control of bacterial infections. Ever since the introduction of antibiotics into clinical use resistance to these antibiotics has emerged sooner or later.
These antibiotic-resistant bacteria have been detected in sewage, and thus there is concern that sewage could be the source of an outbreak of infections caused by these antibiotic-resistant bacteria. There is an opportunity, on the other hand, to use sewage to monitor the prevalence and spread of antibiotic-resistant bacteria within communities or even across the world.
Image Credit: Kateryna Kon/Shutterstock.com
The problem of antibiotic resistance
Starting with penicillin in the 1940s, humanity gained the ability to successfully treat and control bacterial infections. However, the problem of antibiotic resistance has existed as early as the 1950s and now resistance to all available classes of antibiotics has been seen.
This is the case even for antibiotics where resistance was not readily induced in laboratory testing; for example, vancomycin resistance was difficult to induce in laboratory settings, however 7 years after introduction into clinical use vancomycin resistance was reported in coagulate negative staphylococci.
Antibiotic resistance is frequently mediated by genes, which can be transferred between bacteria. The situation is such that the World Health Organization issued a warning that this crisis could become dire. This is not limited to one bacterial species, although there are some which are of more concern than others.
An example of Gram-positive pathogen, methicillin-resistant Staphylococcus aureus (MRSA) infections killed more people in the US than HIV/AIDS, Parkinson’s, emphysema, and homicide combined. Other Gram-positive pathogens of concern include vancomycin-resistant enterococci (VRE), Streptococcus pneumoniae, and multi-drug resistant Mycobacterium tuberculosis.
Gram-negative pathogens are seen as a higher threat than Gram-positive pathogens, as they are developing resistance to virtually all available antibiotics. These notable Gram-negative pathogens include the Enterobacteriaceae family (which include E. coli, Salmonella and Klebsiella species), Pseudomonas aeruginosa, and Acinetobacter species.
Of particular concern is those that produce extended-spectrum beta-lactamases, which renders all beta-lactam antibiotics ineffective.
Can antibiotic-resistant bacteria be spread by sewage?
Antibiotic-resistant bacteria are found in many different environments, including the human gut. Therefore, is there a possibility that antibiotic-resistant bacteria are passed into the sewage system?
A study by Heß and co. looked for the presence of antibiotic-resistant bacteria in airplane sewage. The authors obtained eight sewage samples from five airports, as well as samples from nearby wastewater treatment plants, where it was estimated that 20% of incoming wastewater originated from the airports.
For comparison, the authors also collected samples from six wastewater treatment plants that were not near airports.
The authors used metagenomic sequencing, where the whole extracted DNA is sequenced and then matched to a database of antibiotic resistance genes, as well as quantitative PCR (qPCR) of 14 selected antibiotic resistance genes. The authors also selectively cultured the samples for E. coli as an indicator organism, as it is much known about antibiotic resistance levels associated with E. coli in different regions.
The authors calculated the diversity of antibiotic resistance genes as the number of antibiotic resistance genes found per 1,000 copies of 16S rRNA genes. They found that the diversity of antibiotic resistance genes was significantly higher in airplane sewage compared to untreated sewage from wastewater treatment plants.
Using qPCR data, the authors were able to obtain a relative abundance of 14 antibiotic resistance genes. They found that genes which confer resistance to tetracyclines, aminoglycosides and macrolides were significantly more abundant in airplane sewage compared to wastewater.
When looking at the resistance profile of E. coli isolated from the samples, the authors again found that antibiotic resistance was higher in airplane sewage compared to wastewater; combined resistance to third-generation cephalosporins, fluoroquinolones and aminoglycosides were more frequently seen in E. coli isolated from airplane sewage.
This is concerning, as it demonstrates that antibiotic-resistant bacteria are found more frequently in airplane sewage, and thus can be a source of transmission in multiple parts of the world.

Image Credit: Phatranist Kerddaeng/Shutterstock.com
Can sewage be used to monitor the spread of antibiotic resistance?
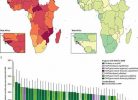
As sewage has been shown to contain antibiotic-resistant bacteria, a study by Hendriksen and co. investigated the possibility of using sewage to investigate the presence of antibiotic-resistant bacteria in urban environments.
The authors collected untreated sewage from 79 sites based in 60 countries. Once DNA was extracted from the sewage, it was sequenced and then matched to a database of antibiotic resistance genes.
When analyzed, they found that the abundance and diversity of antibiotic resistance genes show geographical differences, which was strongly associated with factors such as socioeconomic status, as well as their healthcare systems, and other environmental factors.
This indicates that improving sanitation and strengthening healthcare systems where needed could play an important role in combating the spread of antibiotic-resistant bacteria. This is an example of how sewage was used to determine the burden of antibiotic-resistant bacteria, and the authors propose that this is an ethical way to monitor the spread of these antibiotic-resistant bacteria and predict how the prevalence of these bacteria could change.
Sources
- Ventola, C. L. (2015) The Antibiotic Crisis part 1: Causes and Threats. Pharmacy and Therapeutics 40 (4): 277-283
- van Schaik, W. (2015) The Human Gut Resistome. Phil Trans R Soc B http://dx.doi.org/10.1098/rstb.2014.0087
- Heß, S. et al. (2019) Sewage from Airplanes Exhibits High Abundance and Diversity of Antibiotic Resistance Genes. Environmental Science and Technology DOI: 10.1021/acs.est.9b03236
- Hendrikssen, R. S. et al. (2019) Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nature Communications https://doi.org/10.1038/s41467-019-08853-3
Last Updated: Aug 5, 2020

Written by
Dr. Maho Yokoyama
Dr. Maho Yokoyama is a researcher and science writer. She was awarded her Ph.D. from the University of Bath, UK, following a thesis in the field of Microbiology, where she applied functional genomics toStaphylococcus aureus . During her doctoral studies, Maho collaborated with other academics on several papers and even published some of her own work in peer-reviewed scientific journals. She also presented her work at academic conferences around the world.
Source: Read Full Article