It is essential to face some grim facts about the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pathogen. The ongoing COVID-19 (coronavirus disease 2019) pandemic, started in December 2019, may last for many years. SARS-CoV-2, the causative agent of COVID-19, is the seventh member of the Coronaviridae family known to infect humans.
The SARS-CoV and MERS-CoV (Middle East respiratory syndrome coronavirus), two other members of this family, are the causative agents of the recent outbreaks of the SARS and MERS epidemics. Despite the two-decades-long interaction with these viruses, there is no therapeutic solution against the SARS, COVID-19, and MERS; only supportive care.
In order to develop effective antiviral drugs, scientists design active molecules based on their structures that can prevent or disrupt the specific viral-receptor binding – this is an effective strategy to prevent the virus from entering the host.
To achieve this, Indian researchers from the National Institute of Technology, and MIMS Research Foundation, Calicut, computationally designed a peptide inhibitor that can bind to the receptor-binding domain (RBD) of the spike (S) protein of three viruses – SARS-CoV, SARS-COV-2, and MERS, irrespective of the differences in the type of receptors they bind.
When the peptide binds to the viral receptor, it prevents the binding of the S protein to the respective host receptor, thus inhibiting the entry of these viruses.
“A consensus sequence pattern was derived from these three sequences to obtain the final peptide.”
In this study, the researchers designed a 16-mer peptide (Peptide 7) that binds to the RBD region of SARS-CoV, SARS CoV-2 and MERS-CoV. This work was recently published in the journal PLOS ONE.
Using docking and molecular dynamic simulations, the researchers showed that the peptide-protein (RBD) complex is stable, with a higher binding affinity as compared to the protein (RBD)-receptor (host) complex.
They derived the sequence by taking a sequence common from the three distinct peptides that act on the RBD of the three coronaviruses.
“This peptide was able to interact with the key residues that are involved in binding of the RBDs to their native receptors and thus would be able to prevent the binding and initiation or spread of infection.”
They reported that residues like Lys417 and Tyr449 in SARS-CoV2, Thr486 and Tyr491 in SARS-CoV and Asp510, Arg542, and Glu513 in MERS-CoV RBDs are involved in the interactions of the RBD to their respective receptors.
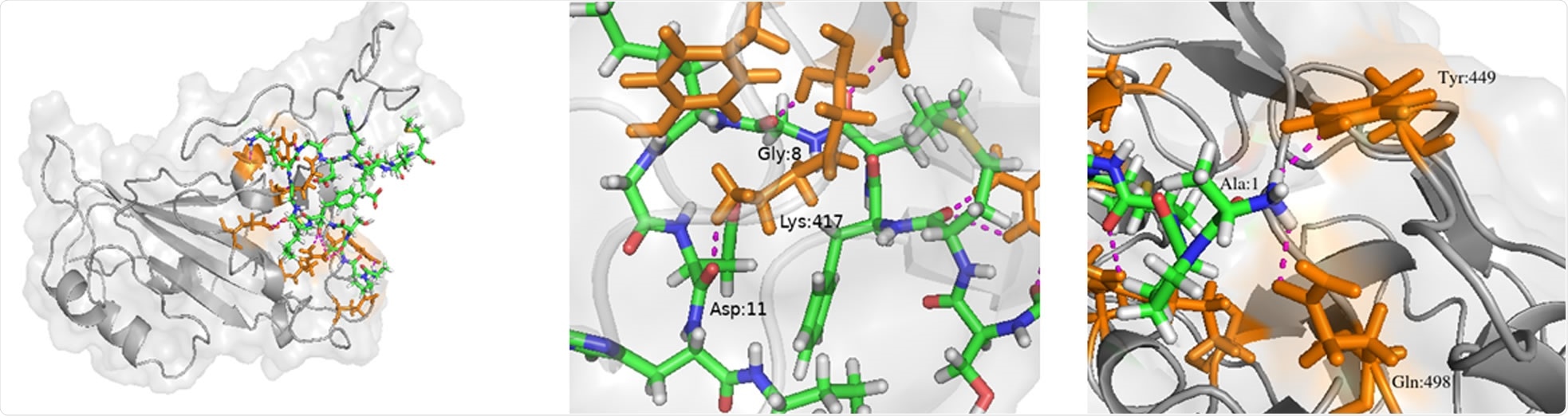
The secondary structure of the Peptide 7 (predicted using Pep2D) consists of a coil, and it did not show any defined secondary structure. The researchers further elaborated that though the RMSD (the measure of the average distance between the atoms of superimposed proteins) is high for the Peptide 7, it robustly prevents the binding of the native receptor. This is attributed to the absence of any secondary structure and the flexibility of the molecule that makes it a good candidate to act on the slightly different RBDs of the three coronaviruses.
The researchers tabulated the binding energy (ΔG) between the native receptor and Peptide 7 with the three coronaviruses. The comparison clearly states the higher affinity of the viral protein with Peptide 7.
In the study, the Radius of gyration and the hydrogen bonds formed in the binding show that the complex of RBD-Peptide 7 is compact and stable during the time of the simulation. However, the researchers also observed some structural fluctuations where there are limited interactions between the peptide and the RBD.
Because of the promising results in this in silico study, the researchers call for further in vitro and in vivo tests to understand the efficacy and the ADMET (absorption, distribution, metabolism, and excretion) properties of Peptide 7, to assess its action as a possible drug candidate.
Few peptide-based drugs are used in the treatment of diabetes, growth deficiency, cancers and some viral diseases. “This peptide could be used as a template and its affinity, stability and bioavailability could be improved by the addition of linker molecules or conjugates such as PEG,” said the researchers.
With an approach of one stone, three birds, this is an important study in the wake of recent epidemic and pandemic outbreaks from coronaviruses. Based on the computational research for designing a viral inhibitor with enormous potential to block the entry of SARS-CoV, SARS-CoV-2 and MERS-CoV, Peptide 7 is the first step. This study may pave the way for the development of peptide-based drugs in the treatment of coronavirus infections.
- V. K. P, Rath SP, Abraham P (2021) Computational designing of a peptide that potentially blocks the entry of SARS-CoV, SARS-CoV-2 and MERS-CoV. PLoS ONE 16(5): e0251913. doi:10.1371/journal.pone.0251913, https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0251913
Posted in: Medical Research News | Disease/Infection News | Pharmaceutical News
Tags: binding affinity, Coronavirus, Coronavirus Disease COVID-19, Diabetes, Drugs, Efficacy, in vitro, in vivo, MERS-CoV, Metabolism, Molecule, Pandemic, Pathogen, Peptides, Protein, Receptor, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Virus

Written by
Dr. Ramya Dwivedi
Ramya has a Ph.D. in Biotechnology from the National Chemical Laboratories (CSIR-NCL), in Pune. Her work consisted of functionalizing nanoparticles with different molecules of biological interest, studying the reaction system and establishing useful applications.
Source: Read Full Article
